Methylation
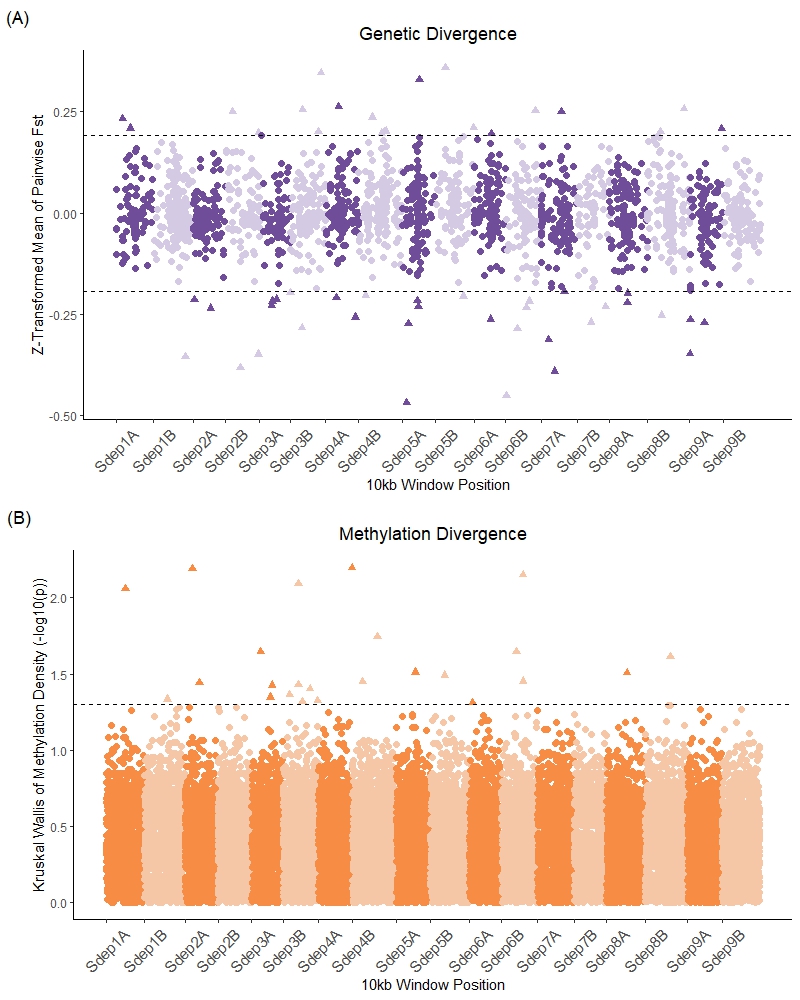
Methylation of CG motifs in many organisms is heritable, epimutable, and contributes to gene expression modulation. In plants, the rate of methylation epimutation is one million times faster than genetic mutation, making it one potential mechanism for rapid responses to the environment. Here we present a cost-effective approach using long-read Nanopore sequencing to simultaneously assess genetic and methylomic divergence and diversity in natural populations and demonstrate its utility in four populations of the the salt marsh plant Salicornia depressa. We find that pairwise genetic kinship between individuals is correlated with methylation profile (ρ = 0.3, p = 0.01). The four populations have similar genetic and methylation diversity across genomic windows, but highly genetically variable windows are uncorrelated with windows with high methylation density variation. Population divergence tests indicate that no genome windows are divergent in both genotype and methylation, as expected statistically if the two divergence types are uncorrelated. Our results suggest that regions of important genetic and methylation variation across populations are physically decoupled. Therefore, studies of complex trait evolution must consider the combined contributions of distinct genetic and methylation polymorphisms. Nanopore sequencing offers a promising avenue to reduce costs of such studies by obtaining genetic and methylation data concurrently. This approach could be a powerful tool to study species that lack standing genetic diversity, including crops and many plants of conservation concern. To encourage adoption and fill the gap of analysis tools available for Nanopore methylation data, we provide custom R scripts used for all methylation analyses.

Genome Assembly
Salicornia depressa (American pickleweed) is the most widespread member of its salt-loving genus in North America. Pickleweeds typically colonize high marsh areas that tides make highly saline. Most other salt marsh plants cannot withstand the same salt pressure, so these bare patches are free from competitive pressures. Understanding the genetic origin of American Pickleweed’s high salinity adaptations has potential application to salt-tolerant agriculture and salt marsh conservation but requires genome resources to study. Salicornia depressa is a tetraploid species, which presents unique challenges to traditional genome assembly pipelines. We present a high-quality, chromosome-scale reference genome of S. depressa and the pipeline we used to assemble it. Our reference is phased to each of the tetraploid’s ancestral subgenomes with a scaffold N50 of 69.3 Mb, BUSCO completeness of 96.4%, and k-mer completeness of 98.4%. The subassemblies are evenly split, with subassembly A containing 56.7% and subassembly B containing 59.1% of genomic k-mers. Our gene annotation identifies 80,883 genes and our methylome annotation contains 2.5 million methylated cytosines. We find that gene and methylation density are negatively correlated across the genome. We also assembled and annotated a chloroplast assembly which includes all expected photosystem, tRNA, and rRNA genes. We provide a guide to our successful assembly pipeline involving >30 programs. Our reference and annotation join resources for three other Salicornia species, allowing global scale ecological-evolutionary studies.
Policy Analysis
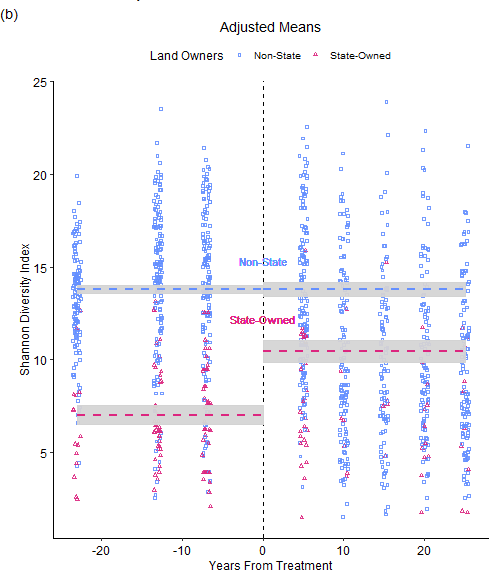
Conservation policymakers use an array of regulatory and legal mechanisms to conserve natural resources on public lands, yet there is no consensus on which mechanisms are most effective at conserving biodiversity. To address this gap, we systematically reviewed papers analysing changes in biodiversity after implementation of state-level conservation policies other than their Endangered Species Act in the United States. Our review reveals a major mismatch between statutory reality and academic literature, with only 12 published assessments of 13 conservation policies and their impacts on biodiversity from over five decades. We present an aggregative meta-summary of the policy mechanisms that have been assessed, the states where they have been implemented, and their impacts. To encourage future research comparing conservation mechanisms, we recommend standardization guidelines, including that policy assessments report the size of area affected and stakeholders involved in implementation of the policy and assess biodiversity changes (e.g. Shannon’s diversity index or population density of focal species) where applicable. As demonstration we present a case study analysis of a 1997 North Carolina amended law governing the state’s forestry council. Using a difference-in-difference method with data from the US Forest Service Forest Inventory and Analysis Database, we find the amendment had a positive effect on Shannon diversity of trees in state forests.